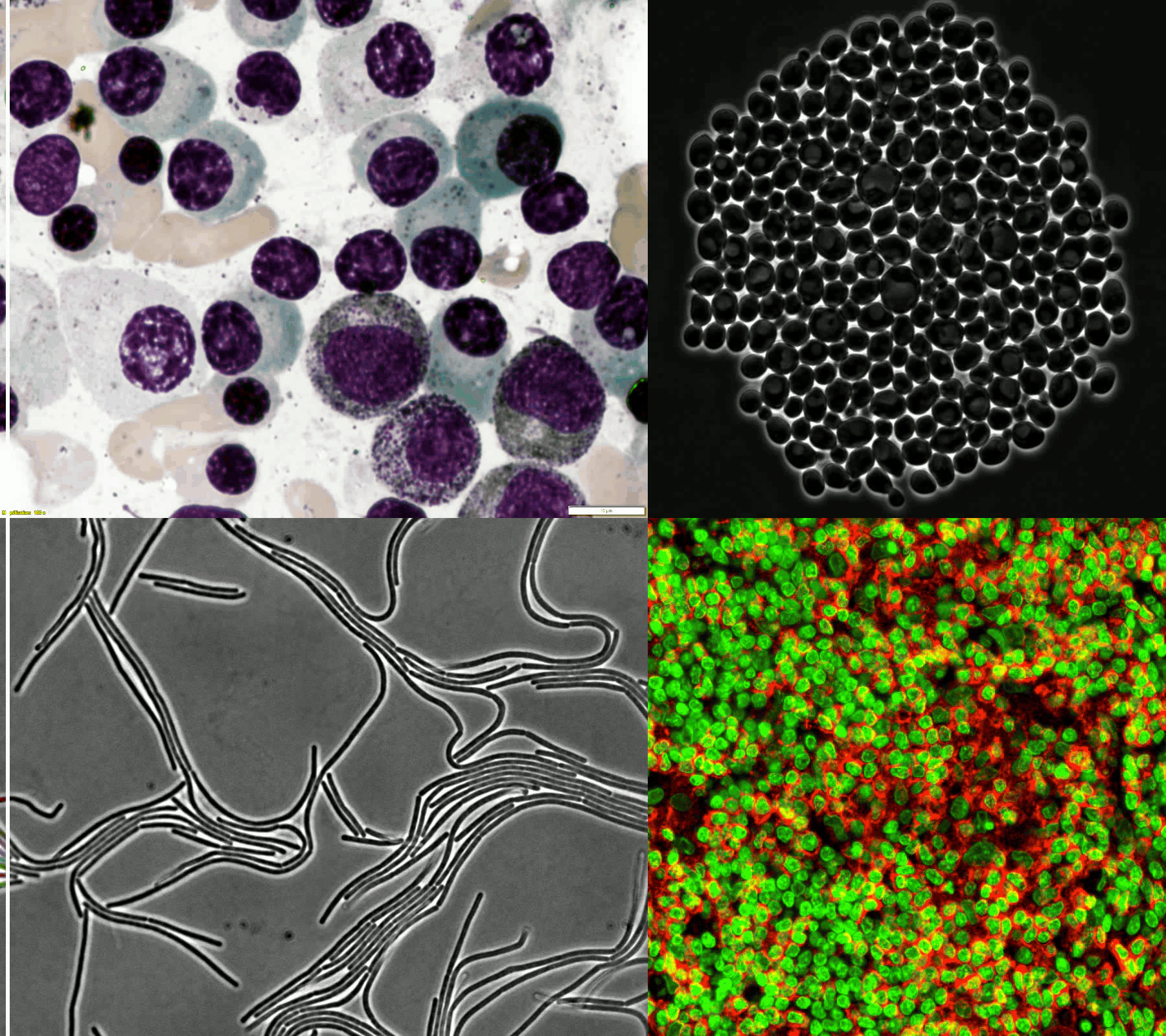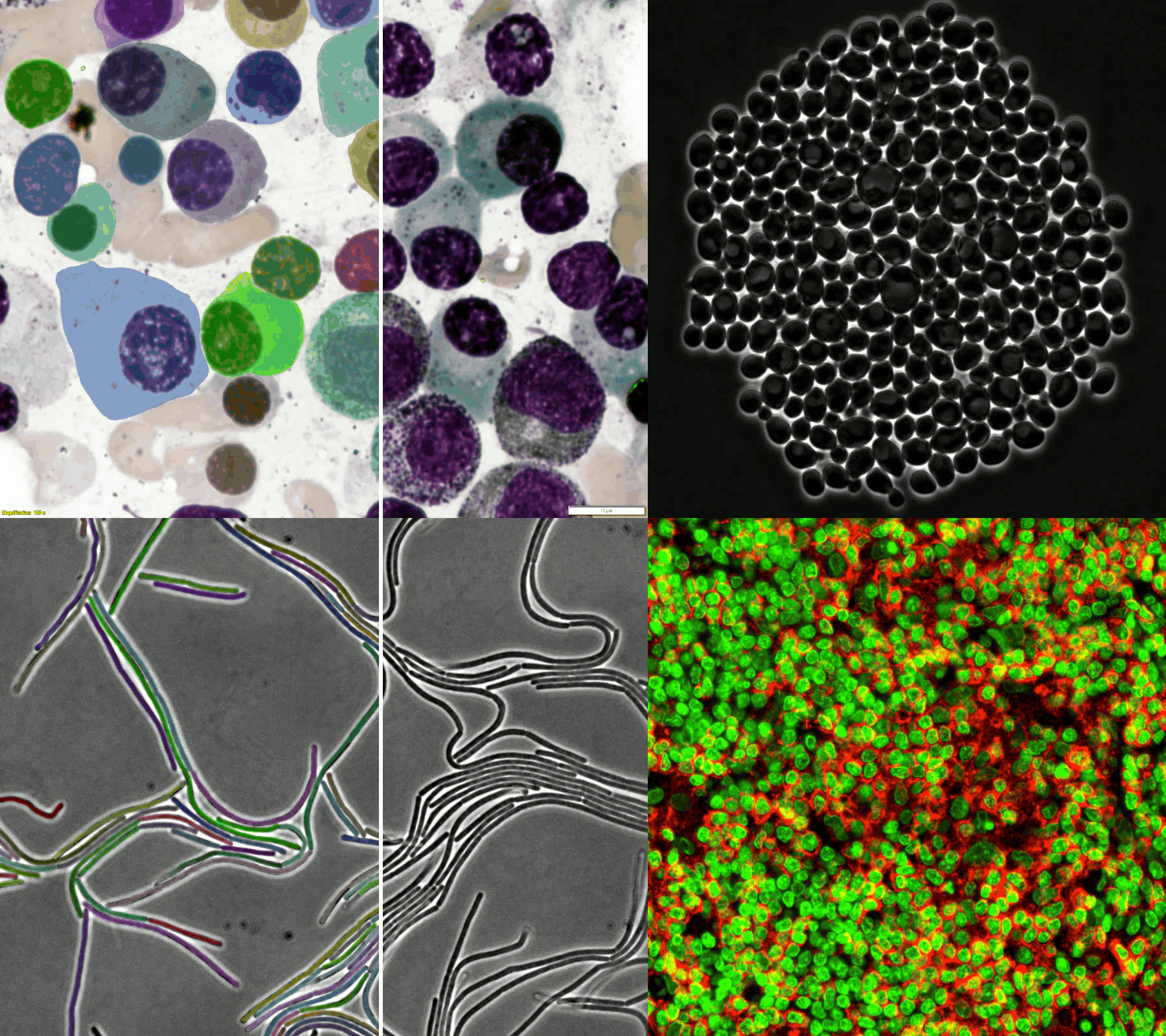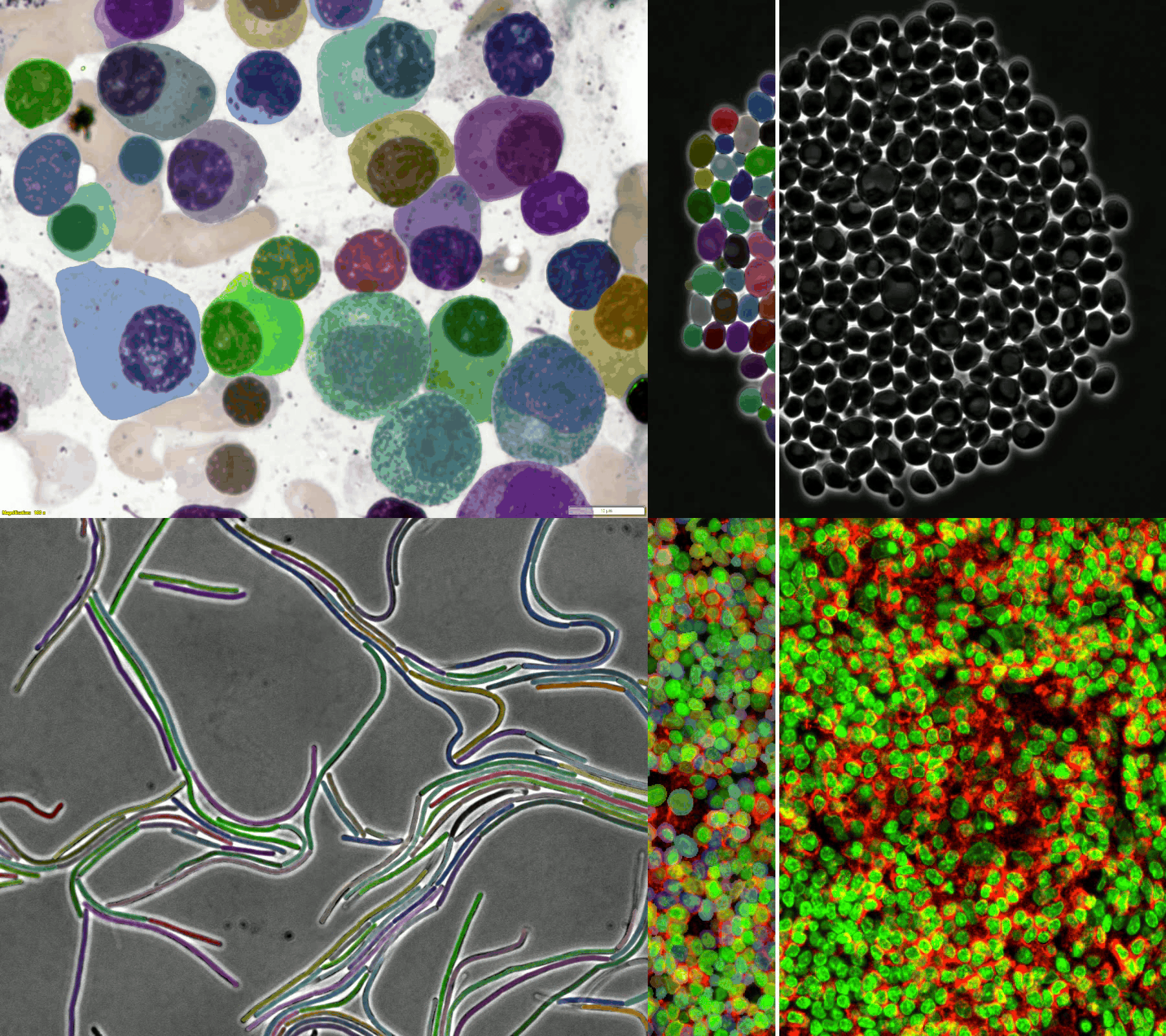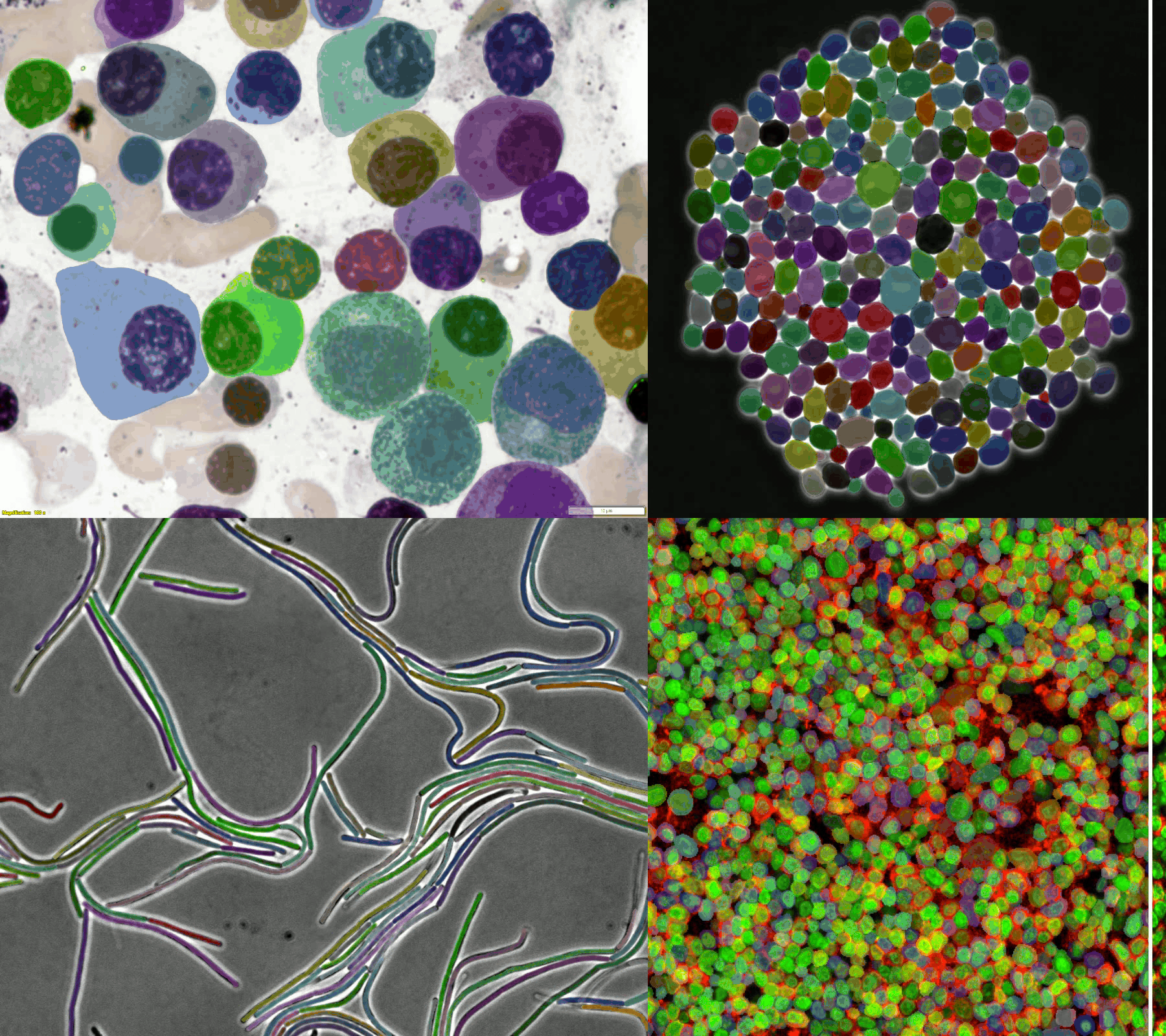
A large and diverse microscopy image dataset
- 50+ different biological experiments
- Four different microscopes modalities
- 900,000+ annotated cells
- Different cell origins, staining methods, morphologies, …
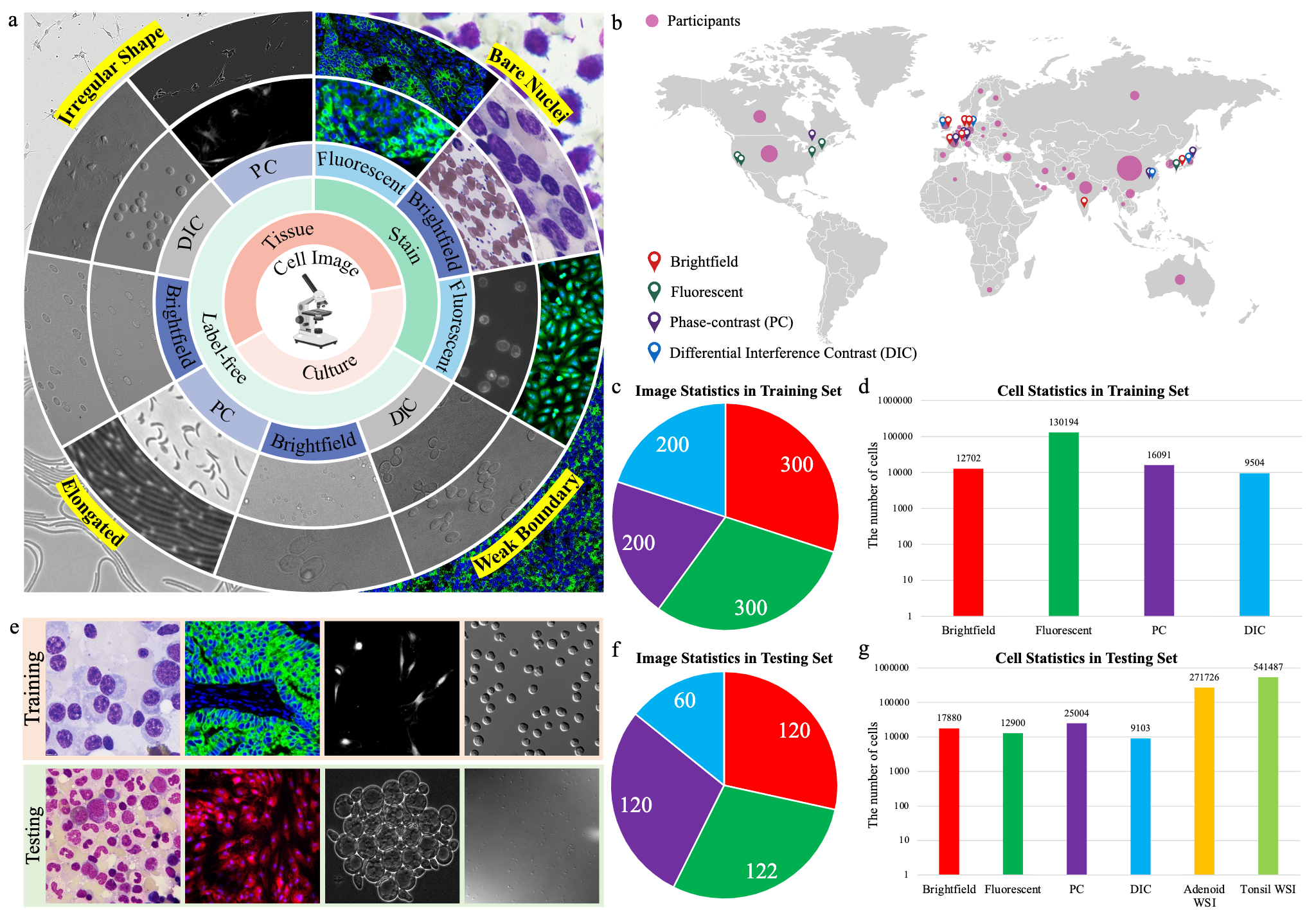
- The challenge provides a diverse microscopy image dataset that includes tissue cells, cultured cells, label-free cells, stained cells, and different microscopes (i.e., brightfield, fluorescent, phase-contrast (PC), and (Differential Interference Contrast) DIC).
- The geographical distribution of data sources and challenge participants. The red, green, purple, and blue address icons denote the countries or regions where the brightfield, fluorescent, phase-contrast, and differential interference contrast image datasets are from, respectively. The size of the pink circle in each country is proportional to the number of participants from the corresponding country.
- The number of images in the training set.
- The number of labeled cells in the training set.
- Randomly selected examples (from left to right: brightfield, fluorescent, PC, and DIC images) from the training set (the 1st row) and testing set (the 2nd row).
- The number of images in the testing set.
- The number of cells in the testing set. There are two fluorescent whole-slide images (WSI) in the testing set.
The top algorithms set new state of the arts
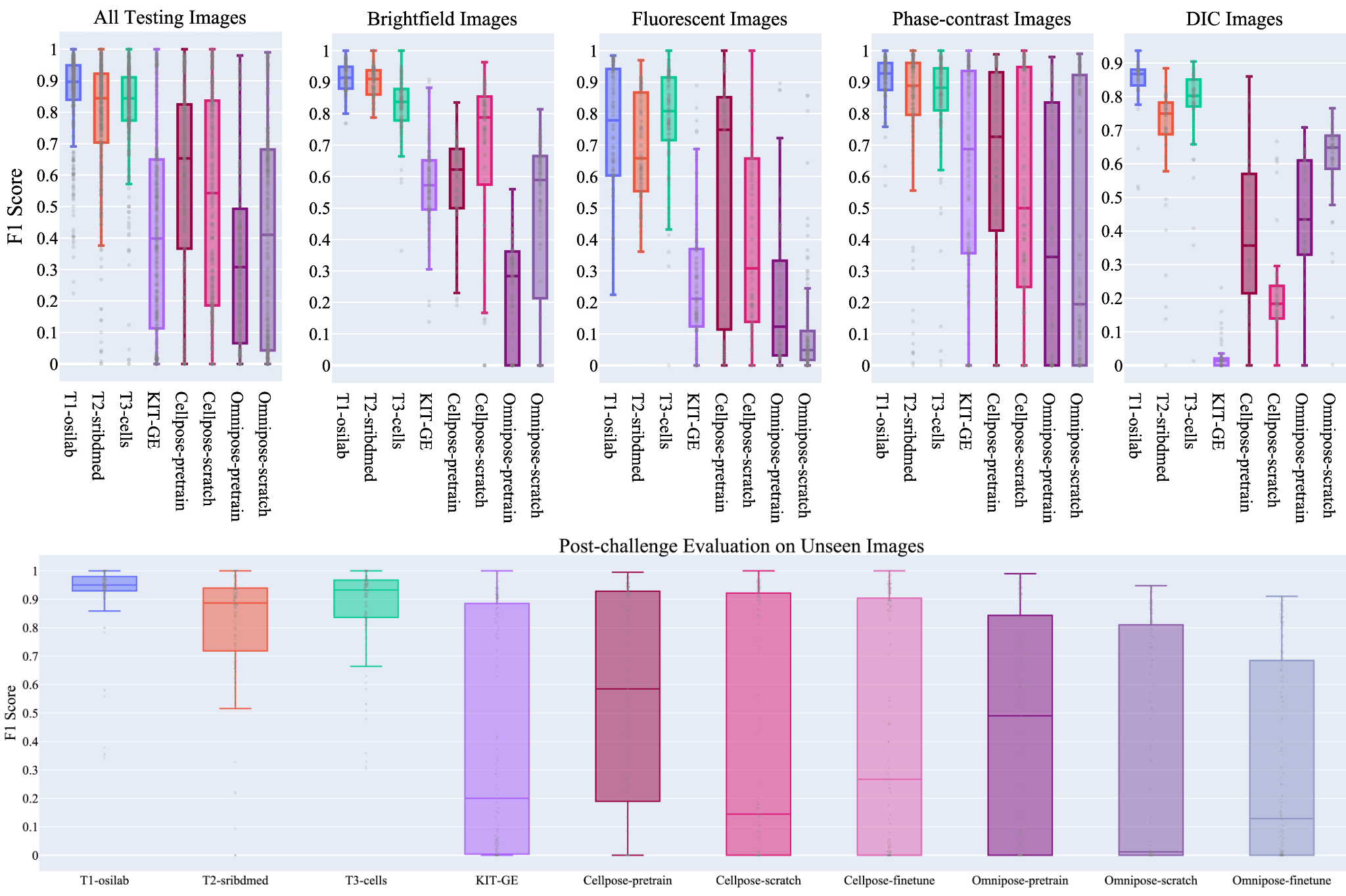
- The winning team, boasting a Transformer-based architecture, has proven to be a remarkable achievement in cell segmentation.
- Top algorithms demonstrated exceptional accuracy and efficiency, surpassing existing state-of-the-art cell segmentation methods such as Cellpose and Omnipose as well as the top algorithm in the CTC challenge.
- All top algorithms have made the code and models publicly available.
| Top Teams | Method | Implementation | Tutorial | Interface | Container |
|---|---|---|---|---|---|
| T1-osilab | Paper | Code | Colab | Napari | Docker |
| T2-sribdmed | Paper | Code | Colab | Napari | Docker |
| T3-cells | Paper | Code | Colab | Napari | Docker |
From SOTA algorithms to biological practice
The top three best-performing algorithms have been integrated into an open-source and user-friendly napari interface. Biological researchers can now conveniently apply these advanced segmentation techniques to their own images without necessitating additional coding.
BibTeX
@article{NeurIPS-CellSeg,
title = {The Multi-modality Cell Segmentation Challenge: Towards Universal Solutions},
author = {Jun Ma and Ronald Xie and Shamini Ayyadhury and Cheng Ge and Anubha Gupta and Ritu Gupta and Song Gu and Yao Zhang and Gihun Lee and Joonkee Kim and Wei Lou and Haofeng Li and Eric Upschulte and Timo Dickscheid and José Guilherme de Almeida and Yixin Wang and Lin Han and Xin Yang and Marco Labagnara and Vojislav Gligorovski and Maxime Scheder and Sahand Jamal Rahi and Carly Kempster and Alice Pollitt and Leon Espinosa and Tâm Mignot and Jan Moritz Middeke and Jan-Niklas Eckardt and Wangkai Li and Zhaoyang Li and Xiaochen Cai and Bizhe Bai and Noah F. Greenwald and David Van Valen and Erin Weisbart and Beth A. Cimini and Trevor Cheung and Oscar Brück and Gary D. Bader and Bo Wang},
journal = {Nature Methods},
year = {2024},
doi = {https://doi.org/10.1038/s41592-024-02233-6}
}
This website template is borrowed from here. Thank you!